I actually am still here, but Spring 2007 finals went straight into Summer 2007 classes, and it’s pretty intense. A six-week Immunology and Immunology lab alongside an entire semester of “Speech” class crammed into 4 weeks means I’m pretty frantic at the moment.
Although the semester is over and the paper turned in, I am still working on my 8 beer-bacteria isolates. In fact, re-doing “colony” PCR with a few µl of broth culture seems to have actually worked pretty well:
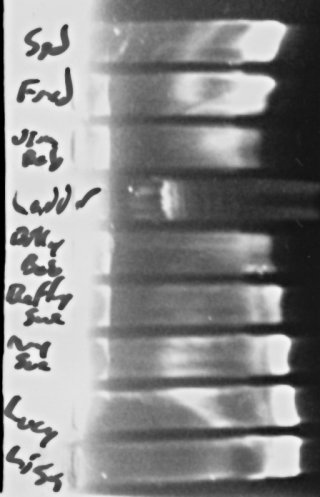 To the left, there, you can see the results. Interestingly, if you look just to the right of where the “wells” are in the gel, in several lanes you can make out a visible band of obviously large DNA molecules, which I presume are genomic DNA from the bacteria. The fact that they seem brighter in the lanes where the 16s band at the end (or at least, I’m ASSUMING that’s what that is, and my reaction worked) is dimmer tends to support my suspicion that I’m “swamping” my reaction with the template DNA and making it hard for the reaction to work sometimes.
To the left, there, you can see the results. Interestingly, if you look just to the right of where the “wells” are in the gel, in several lanes you can make out a visible band of obviously large DNA molecules, which I presume are genomic DNA from the bacteria. The fact that they seem brighter in the lanes where the 16s band at the end (or at least, I’m ASSUMING that’s what that is, and my reaction worked) is dimmer tends to support my suspicion that I’m “swamping” my reaction with the template DNA and making it hard for the reaction to work sometimes.
Anyway, once I’ve worked out how to actually do a useful Restriction Enzyme digest to get an idea of how many of my isolates are really unique, I should hopefully be about ready to get them sequenced so that I can identify them. Then, I’ll be ready to try them out on my thesis project. Meanwhile, both my current DNA extraction from my recent thesis project sample and the amplified 16s gene bands for my lactic-acid bacteria from the beer are sitting in the freezer.
I’ve got an intense couple of days now, but hopefully I’ll have time to blog a bit more over the weekend.