As you can see, I’ve got DNA. I’ve been trying to get this stuff successfully extracted and the 16s rDNA amplified for months (off and on) now. Looks like doing the whole-genome-amplification step first did the trick – this is from a set of mixed halophiles in a phlogisticated environment growing in approximately 18% salt solution, and they grow very slowly. It’s hard to get enough DNA extracted from such a small population to do useful work with.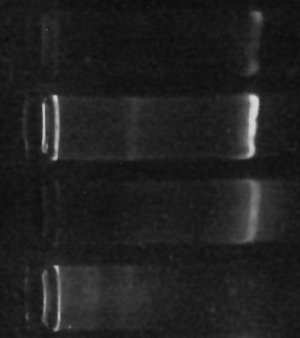
The gel “bands” you see to the right of the image are (or at least should be) made of copies of the DNA which codes for various “16s small-subunit ribosomal RNA” sequences for the one-or-more different kinds of prokaryotes living in my culture. The brighter the band, the more DNA is there.
Since all of the samples were processed exactly the same way, then, the brightness of the band should, at least indirectly, indicate how many bacteria were in each sample to begin with. This isn’t necessarily true – there can be variation in how many copies of the gene each kind of bacteria has, so if the populations are very different the results could be misleading. Still, it’s gratifying that my little ‘proof-of-concept’ experiment not only finally gave me some DNA but even shows exactly the kind of difference I originally hoped for. (The second “lane” from the top with the brightest band was SUPPOSED to be enriched for certain types of bacteria, according to my hypothesis. The first “lane” should have had less, and it does. The third lane is my “positive control”, growing without special influences, and the fourth lane with no DNA visible is my negative control, which I hoped would have little or no DNA (indicating little or no bacteria growing in it) – and that’s what I see.
It doesn’t prove anything at this point, but finally getting results and having them turn out to look the way I’d hoped is a good start. I wonder if I can get them into a clone library, separated, and sequenced before next weekend?
I’ll have to remember to thank last semester’s “Senior Seminar in Microbiology” instructor for assigning me that paper[1] – I thought some of the technology described in it sounded like it’d be useful to me personally.
Anybody else going to the Northwest Regional ASM meeting next weekend?…
[1] Wu L, Liu X, Schadt CW, Zhou J: “Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification.” Appl Environ Microbiol. 2006 Jul;72(7):4931-41.